AMBER force field calculation
This section describes how to perform a structure optimization and vibrational analysis using AMBER force field. We use TRP Cage as an example.
[Structure optimization and vibrational analysis]
First, prepare molecular structure data of TRP Cage in Amber prmtop format and Amber inpcrd format as trpcage.prmtop and trpcage.crd.
For creating those files, use LEaP program in AmberTools. About how to use it in detail, please refer to Amber manual and tutorial.
The amino acid sequence of TRP Cage is [NLYIQWLKDGGPSSGRPPPS] in one letter code. According to this, create the LEap input file containing below (file name: tleap.in).
source leaprc.protein.ff14SB
TRP = sequence { NASN LEU TYR ILE GLN TRP LEU LYS ASP GLY GLY PRO SER SER GLY ARG PRO PRO PRO CSER }
saveAmberParm TRP trpcage.prmtop trpcage.crd
quit
After that, execute tLEap by the below command. The trpcage.prmtop and trpcage.crd will be generated. Note that you should set up an environment setting for AMBER program in advance.
tleap -f tleap.inenter
You can find the trpcage.prmtop and trpcage.crd in Sample_Files folder in the folder installed CONFLEX (Sample_Files\CONFLEX\amber\trpcage.prmtop.[prmtop|crd]).
About file formats of prmtop and crd files, please refer to Amber file formats. Regarding the prmtop file in detail, please also refer to prmtop.pdf.
The first part of contents in the trpcage.prmtop file is shown below. There are the number of atoms, the number of residues, and atomic name assigned to each atom, and so on. The information in the trpcage.prmtop file is not changed in the simulation.
%VERSION VERSION_STAMP = V0001.000 DATE = 12/01/20 16:04:37
%FLAG TITLE
%FORMAT(20a4)
NASN
%FLAG POINTERS
%FORMAT(10I8)
304 12 150 160 346 219 700 656 0 0
1701 20 160 219 656 53 124 138 26 0
0 0 0 0 0 0 0 0 24 0
0
%FLAG ATOM_NAME
%FORMAT(20a4)
N H1 H2 H3 CA HA CB HB2 HB3 CG OD1 ND2 HD21HD22C O N H CA HA
CB HB2 HB3 CG HG CD1 HD11HD12HD13CD2 HD21HD22HD23C O N H CA HA CB
HB2 HB3 CG CD1 HD1 CE1 HE1 CZ OH HH CE2 HE2 CD2 HD2 C O N H CA HA
CB HB CG2 HG21HG22HG23CG1 HG12HG13CD1 HD11HD12HD13C O N H CA HA CB
HB2 HB3 CG HG2 HG3 CD OE1 NE2 HE21HE22C O N H CA HA CB HB2 HB3 CG
〜〜〜〜〜〜〜〜〜〜
The first part of contents in the trpcage.crd file is shown below. There are name assigned to the molecule, the number of atoms, and x, y, and z coordinates of each atom.
NASN 304 3.3257700 1.5479090 -0.0000016 4.0461540 0.8399910 -0.0000029 2.8230940 1.4995080 -0.8746870 2.8230970 1.4995070 0.8746850 3.9700480 2.8457950 -0.0000001 3.6716630 3.4001290 -0.8898200 3.5769650 3.6538380 1.2321430 2.4969950 3.8010750 1.2413790 3.8774840 3.1157950 2.1311970 4.2537000 5.0171120 1.2321440 5.0052990 5.3404060 0.3150720 3.9848850 5.8179090 2.2659170 4.4080150 6.7337020 2.3147430 3.3596110 5.5042970 2.9944640 5.4855410 2.7052070 -0.0000044 6.0088240 1.5931750 -0.0000084 〜〜〜〜〜〜〜〜〜〜
[Execution by Interface]
First, open the trpcage.prmtop file by CONFLEX Interface. When opening the file, select [AMBER prmtop file] as the file format. Since there is no coordinate data in the prmtop file, the dialog will be displayed once again when clicking . In the dialog displayed, select the trpcage.crd that has the coordinate data and click .
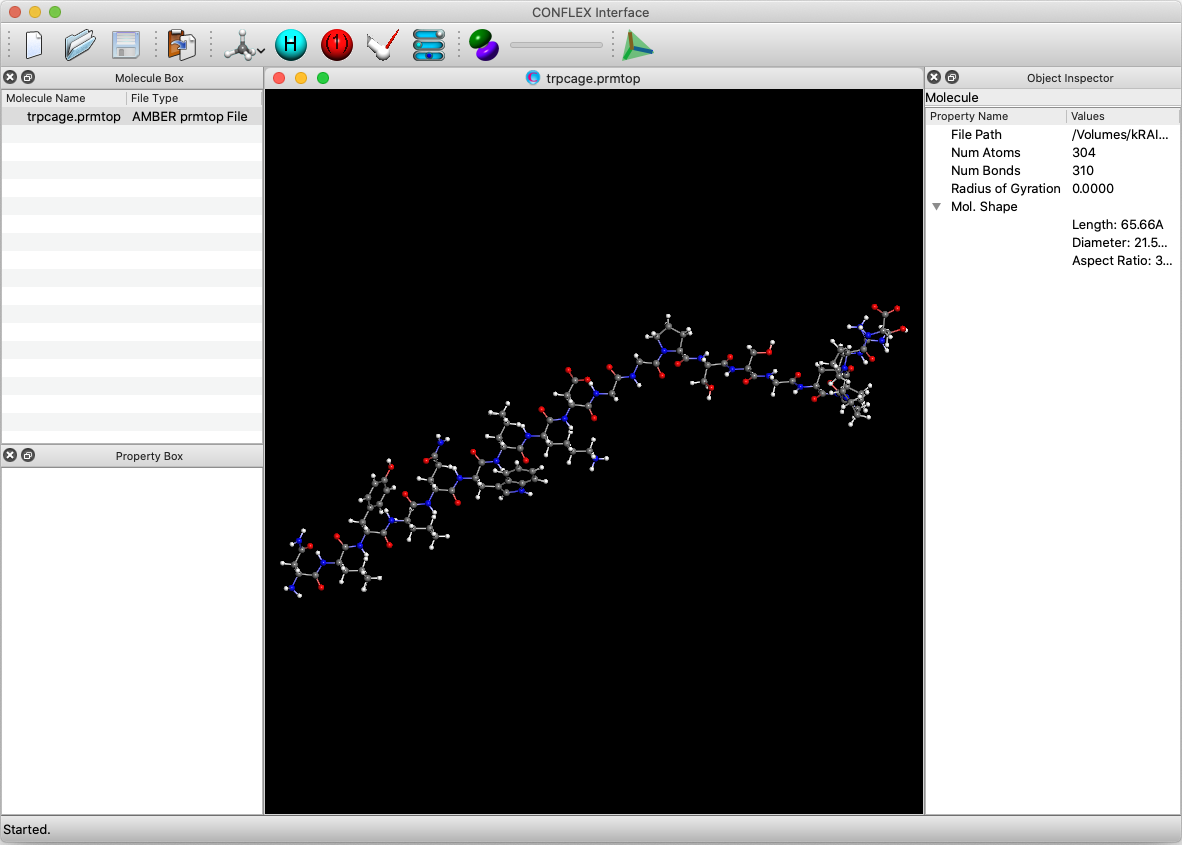
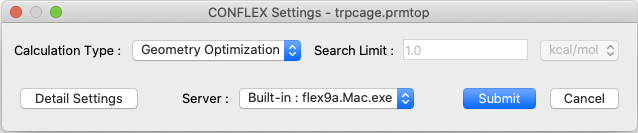
Select [CONFLEX] in Calculation menu, and click in the calculation setting dialog displayed. The calculation of molecular structure optimization and normal mode analysis will be performed. When the molecular file is in Amber format, the Interface automatically select Amber force field.
[Execution by command line]
Store the two files of trpcage.prmtop and trpcage.crd in an one folder, and execute below command. The calculation will start.
C:\CONFLEX\bin\flex9a_win_x64.exe -par C:\CONFLEX\par trpcageenter
The above command is for Windows OS. For the other OS, please refer to [How to execute CONFLEX].
[Output files in the structure optimization]
After the calculation finished, mainly below files are outputted.
- trpcage.bso
- This file has force field parameters were used the calculation, energies of each interaction as well as thermodynamic quantities, frequencies, and vibrational modes obtained by the normal mode analysis.
- trpcage-F.mol2
- This files has the optimized structure in Sybyl-Mol2 format.
- trpcage-F.prmtop
- The contents of this file match the trpcage.prmtop file.
- trpcage-F.crd
- This files has the optimized structure in AMBER Inpcrd format.
[If you executed the calculation by using command line]
You can find trpcage.bso file in the folder storing the input files. You can visualize the optimized structure by opening the bso file by CONFLEX Interface.