Crystal structure optimization using cif file
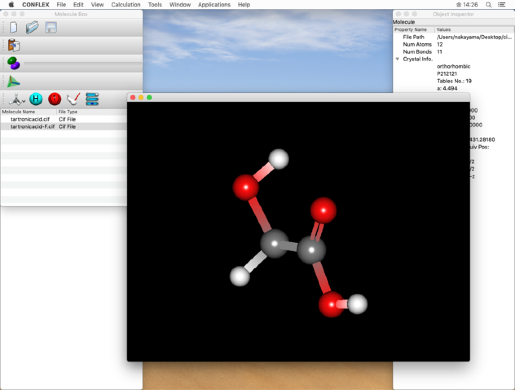
In this section, we will use the "tartronicacid.cif" file in the following folder as an example:
CONFLEX/Sample_Files/CONFLEX/crystal/optimization/cif_file
Please copy this file to an appropriate location under your home directory and work on it.
- Click "Open" from the "File" menu to open the copied "tartronicacid.cif" file.
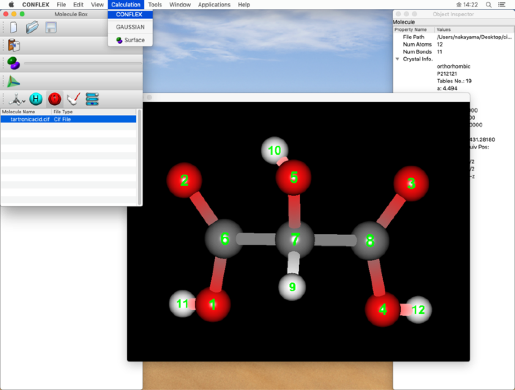
- Select "CONFLEX" from the "Calculation" menu to display the CONFLEX Settings dialog.
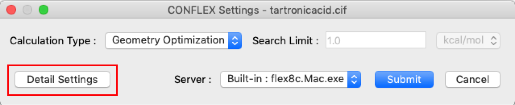
- Click "Detail Settings" in the CONFLEX Settings dialog to display the Detail Settings dialog.
- First, set the type of calculation.
- Next, set the optimization method.
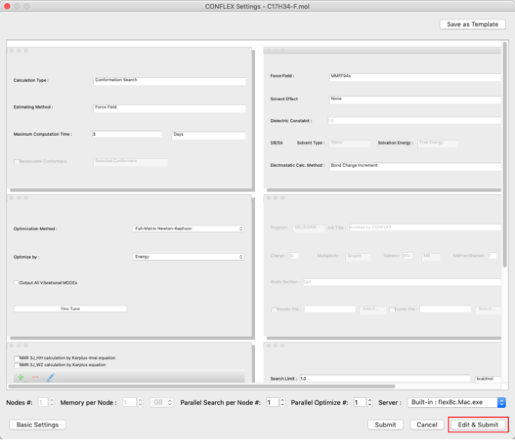
- Click "Edit & Submit" in the Detail Settings dialog.
- The bond order of the molecule is set by the "CIF_BOND=" keyword.
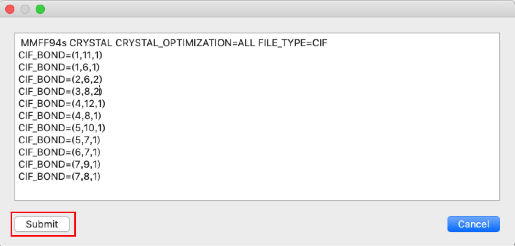
- Make sure the bond orders are set correctly, and click "Submit". .
- When the job is started, the "Job Manager" will be displayed.
-
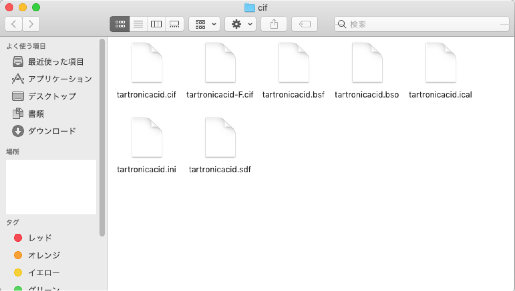
The folder contains "tartronicacid.cif" and six other files:
- tartronicacid.bsf
- tartronicacid.bso
- tartronicacid.ical
- tartronicacid.ini
- tartronicacid.sdf
- tartronicacid-F.cif
When opening a file, please select "Cif File (*.cif *.cmf)" as the file type.
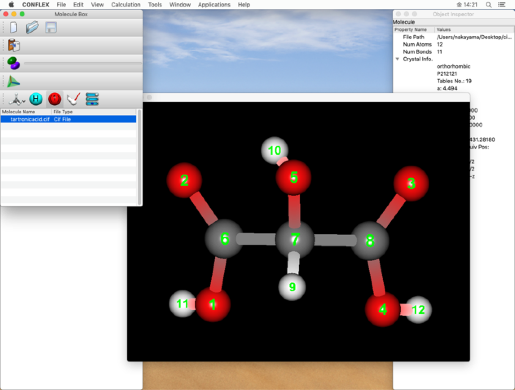
In the step below, we will set the bond order of the double bond, so please check the atomic number here.
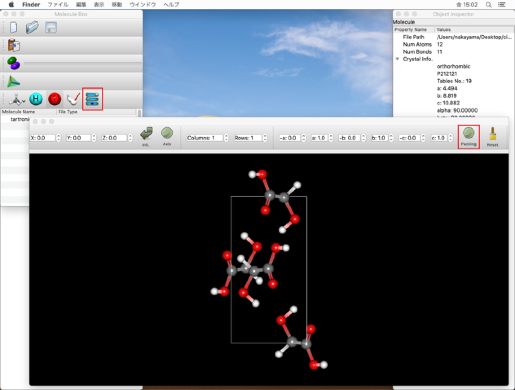
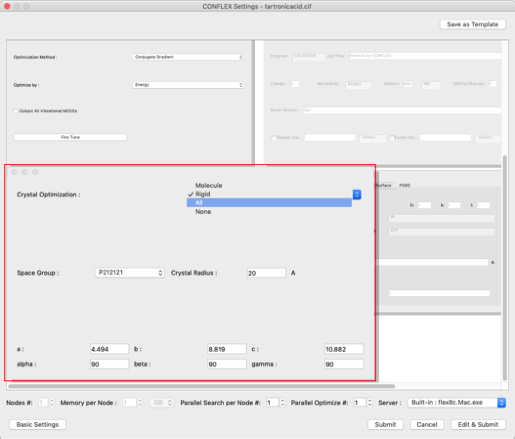
Select "Molecular Crystal" from the "Calculation Type:" pull-down menu in the "General Settings" dialog on the top left.
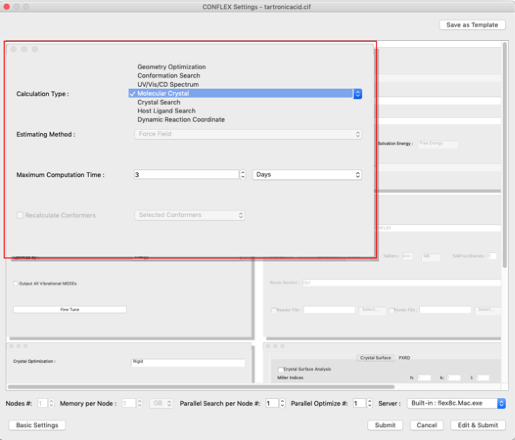
Select "ALL" from the "Crystal Optimization:" pull-down menu in the "Crystal Optimization" dialog on the lower left. The values in the cif file will be displayed for the lattice constants and space groups.
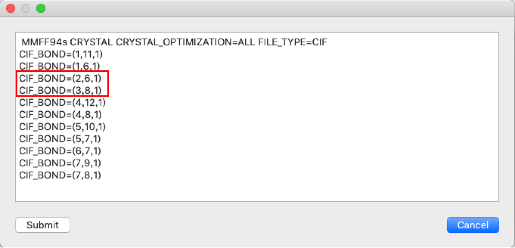
Bonds between atoms 2-6 and 3-8 of the input molecule are double bonds, so change the 1 in "CIF_BOND=" to "2"
CIF_BOND=(2,6,1)
CIF_BOND=(3,8,1)
↓
CIF_BOND=(2,6,2)
CIF_BOND=(3,8,2)
The serial number of the atoms are checked in the molecule window.
The crystal structure optimization job will be started.
After confirming that the "State" is "Finished", double-click on the row indicated by the red frame.
Double-click to open the molecule window and view the optimized structure (tartronicacid-F.cif).