Flow of Calculations
In this chapter, we will discuss the various functions in CONFLEX and how to configure them in the CONFLEX Interface. We will also explain how to run CONFLEX from the command line.
Please note: Since there is no function to create or draw structures in CONFLEX, a third party chemical drawing application is required. Files will need to be saved in the MDL Mol or Sybyl mol2 file format.ここは、CONFLEXを用いて配座解析を行う流れを説明します。
Opening Files
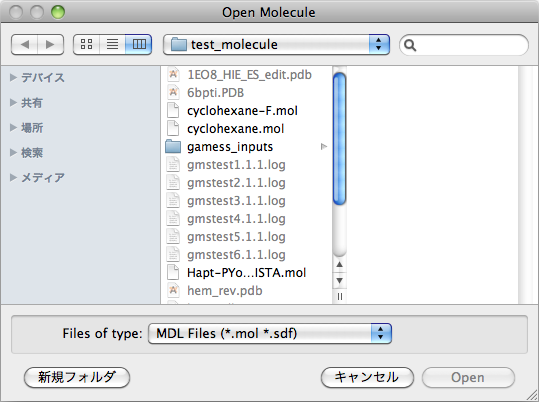
When [Open] is selected from the [File] menu, the following dialogs are displayed.
A file format can be designated using the pop-up at the bottom of the dialog.
When a molecular structure file is selected and the [Open] button is clicked, the file opens and the structure is displayed in the graphic window.
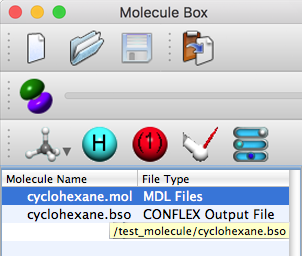
When a file is opened, a list is displayed in the [Molecule Box] dialog.
Hover the mouse cursor over the file name a few seconds, the file path is displayed.
After an item in the list is right-clicked and [Open Folder] is selected from the pop-up menu, the actual file is displayed.
Starting CONFLEX Calculations
In starting calculations, [CONFLEX] is selected from the [Calculation] menu. When this is selected, the following dialogs are displayed.
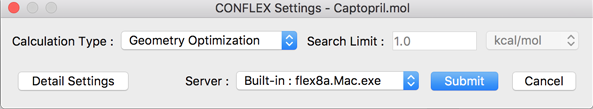
The contents of the dialog are described below:
- Calculation Type:
- The calculation options are selected here:
- Geometry Optimization
- Conformation Search
- NMR J coupling
- Search Limit:
- The search parameters are designated here using the either kcal/mol or percentage (%).
- Server:
- This is used to designate the location of the CONFLEX Engine, i.e. where calculations are to be carried out. This can be the local computer or a remote server. Servers can be added or deleted in the [Preferences] settings.
- Detail Settings
- This is used to toggle on various displays in CONFLEX. See the [Detail Settings for CONFLEX Calculations] on this manual.
- Submit
- This is the calculation start button. Clicking it submits the job to the Job Manager.
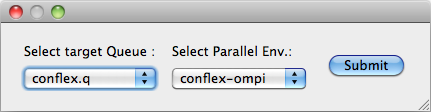
If the compute server is a remote server on which Grid Engine, Platform LSF, or PBS is installed, the dialog box to select the queue name and parallel environment will appear. In the case of Platform LSF or PBS, the name of the queue will be displayed. Select the appropriate one and click the [Submit] button.
Job Manager
When the [Submit] button for the calculations is clicked, the job is registered in the Job Manager where it is queued for execution. The job is executed depending on the CPU load.
To display it manually, select [Job Manager] in the [Tools] menu.
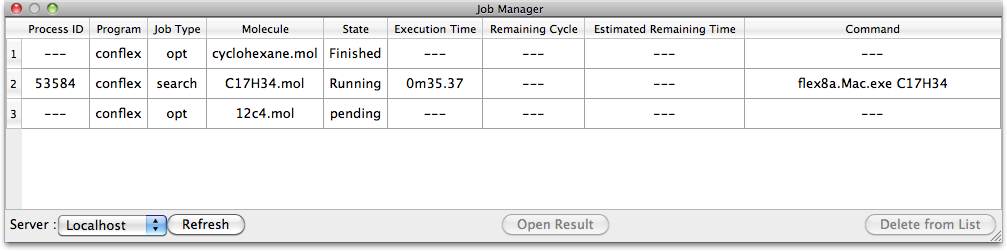
The process for each job is checked every second, or as designated in the [Preferences] settings, and the display is updated. To refresh the results display when an automatic check has not been designated, click the [Refresh] button at the lower left of the dialog.
[Server: Localhost] indicates the computer which is running the CONFLEX Engine. Details for each item are as follows:
- Process ID
- This is the identification number of the job being executed.
- Program
- The name of the job being executed is displayed here.
- Job Type
- The execution contents of the job are displayed in simple form.
- Molecule
- The file name of a molecule being calculated is displayed here.
- State
-
The execution status of a program is displayed here.
There are three states: Pending, Running, and Finished. - Execution Time
- The time elapsed from the start of execution is displayed here.
- Remaining Cycle
- When conformational search calculations are being carried out using CONFLEX, the number of remaining cycles is displayed.
- Estimated Remaining Time
- The estimated time to completion is displayed. This is based on the time elapsed for each cycle being executed. However, there may be wide variations as the calculations proceed.
- Command
- The execution command is displayed here.
Canceling a Job
A job which has been submitted in error can be cancelled. To do so, select the job and click the [Kill Job] button that appears in the lower right of the dialog.
Deleting Calculation History
All of the past jobs are stored as history in the [Job Manager], however, they may be deleted when they are no longer needed. This only applies to jobs in the [Pending] or [Finished] state. To do so, select the job and then click the [Delete from List] button that appears in the lower right of the dialog.
Display of Calculation Results
To review the results of a finished job, double-click it in the list of Job Manager, or select it and click [Open Result]. In a job on an external server case, output files are copied via a network to the local computer then opens an output file.
- For CONFLEX optimization calculations
- The CONFLEX calculation log (*.bso) file opens.
- For CONFLEX conformational search
- The conformation list stored in MDL SD format (*.sdf ) is opened.
- For CONFLEX crystal search
- Opens a list of crystal polymorphs saved in CIF format.
- For Gaussian calculations
- The Formatted Checkpoint file (*.fchk) opens.
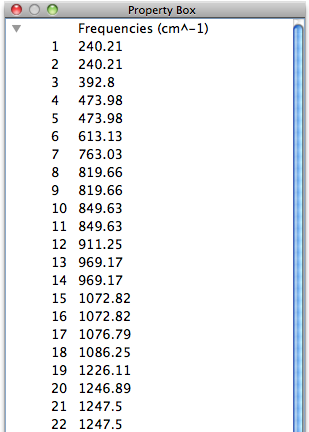
Normal Vibration
When [Vibration] in the [View] menu is selected, the normal mode vectors are displayed. When an object is clicked and selected from the list of vibrations listed in the [Property Box], it is combined with the current display.
Vibrations that do not contain vector data in the file will be grayed out and cannot be selected.
When [Controller] in the [View] menu is selected while a vector is displayed, a slider which adjusts the length of the vector is displayed inside the window.
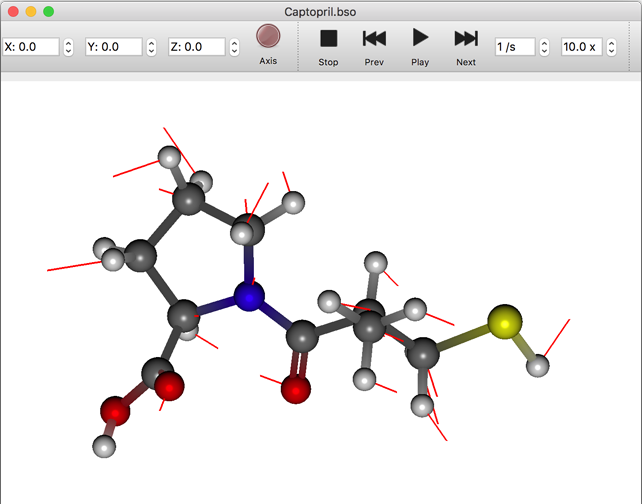
Conformation List
When opening an MDL SD file to review the results of a conformational search, the most stable structure is displayed in the graphic window and an overview of the conformation is displayed in the [Property Box]. When a structure in the list is selected, the structure inside the graphic window is changed to that conformation.
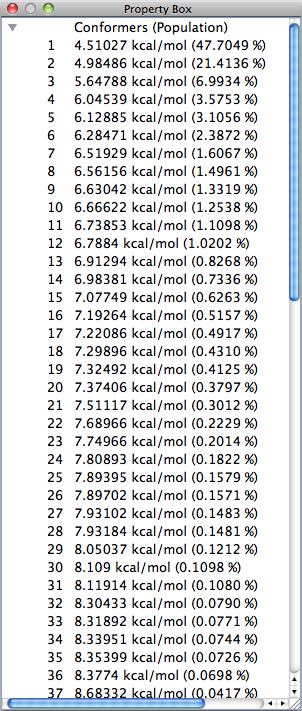
The numerical value displayed by [kcal/mol] on each line is the energy for each conformation and the numerical value in parentheses is the probability of existence of the conformation based on the Boltzmann distribution.
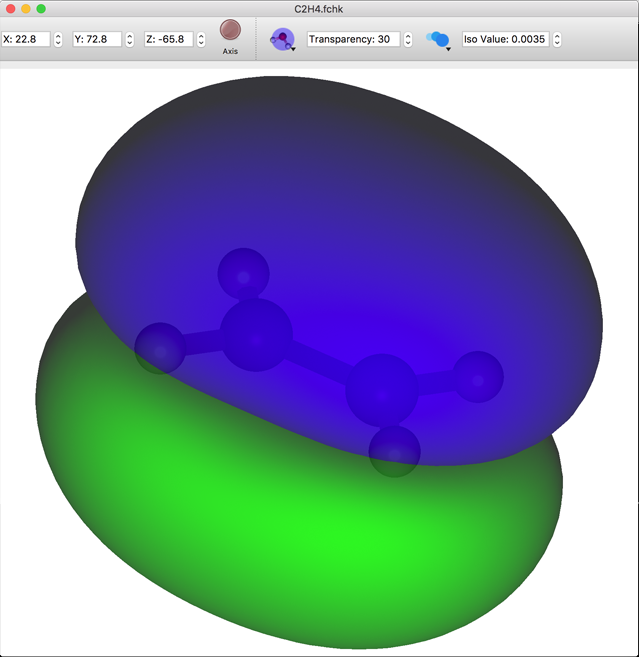
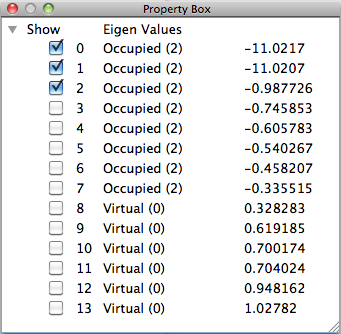
MO List
When a Gaussian Formatted Checkpoint file is opened, an overview of the molecular orbital energies are displayed in the [Property Box]. The respective check boxes are displayed in the items in the list. After ticking an item, when [MO] is selected from the [Calculation] menu or when the [MO] button of the toolbar is clicked, the display data to be checked are calculated. Then, the surface expressing the molecular orbital is displayed in the graphic window. When another checked item is clicked, the MO display switches to that item. When other unchecked orbitals are displayed, the abovementioned operation should be repeated.
The size of the surface displayed is in accordance with the [Preferences] setting, however, the size can be changed in real time using the [Controller]. When the [Controller] is selected from the [View] menu, or the [Controller] button on the tool bar is clicked, the controller can be used to adjust the size of the surface as well as the transparency level in the molecular display window.